Proteasomes and the Ubiquitin-Proteasome System (UPS)
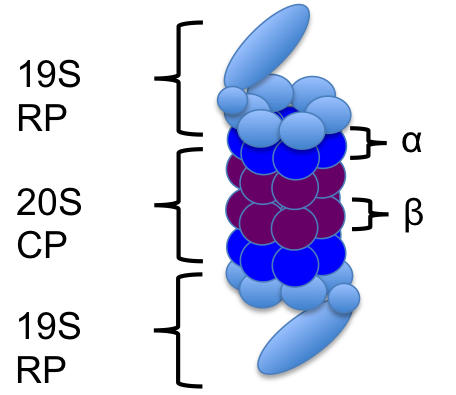
The 26S proteasome is a protein complex that is responsible for the degradation of proteins that are misfolded, damaged or no longer being used. It's structure consists of the 19S regulatory cap(s) and the 20S core particle. The 19S cap is the part of the proteasome that recognizes the proteins that have been tagged for degradation. The protein "tag" is a polyubiquitin chain at least 4 ubiquitin long. (Ubiquitin is a 76 amino acid residue which when tagging a protein for degradation binds to the other ubiquitin at the lysine-48 sites only.) Once recognized by the proteasome, the tagged protein starts to go through the proteasome. The 19S cap unfolds the protein and the ubquitin chain is deubiquitinated - the ubiquitin are now free to be reused in the cell. The 20S core particle is where all of the degradation machinery is contained, and it is here that the protein is led into where it can be broken down. The end result is a proteasome that is ready to be used again, and peptides. The amino acids can be reused later for protein synthesis.
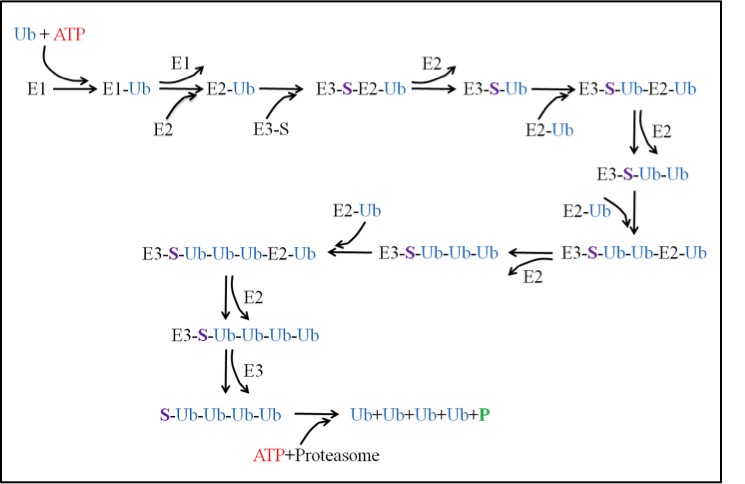
The ubiquitin-proteasome system (UPS) is then a pathway in the cell that is responsible for degradation of proteins. The pathway can be broken up into two main processes: ubiquitination and degradation. The ubiquitination process requires three enzymes: E1, E2 and E3. The activating enzyme E1 uses an ATP and activates a ubiquitin. The activated ubiquitin is then ready for the conjugating enzyme E2. E2 takes the ubiquitin and is now ready for the ligase E3. E3 has the important role of binding a substrate that needs to be degraded. Once bound to the substrate, E3 will then accept the E2-Ub complex and the ubiquitin can then be transferred from the E2 to the substrate. The process repeats itself until the substrate acquires a chain of ubiquitin at least 4 ubiquitin long. At this point, the tagged substrate can then be released from E3 and recognized by the proteasome. This takes us to the degradation process described above.
In vivo single molecule measurements using STED microscopy
We will use STED microscopy to study the life cycle of a protein from birth at the ribosome to death at the proteasome in yeast cells (Saccharomyces cerevisiae). Using GFP as our substrate, we will be able to monitor the formation of the protein in the cell as it will correlate to an increase in fluorescence, and likewise, we will be able to monitor the death of the protein as it will correlate to a decrease in fluorescence. We plan to measure degradation rates and use the data to elaborate simple models like the one below to include temporal behavior.
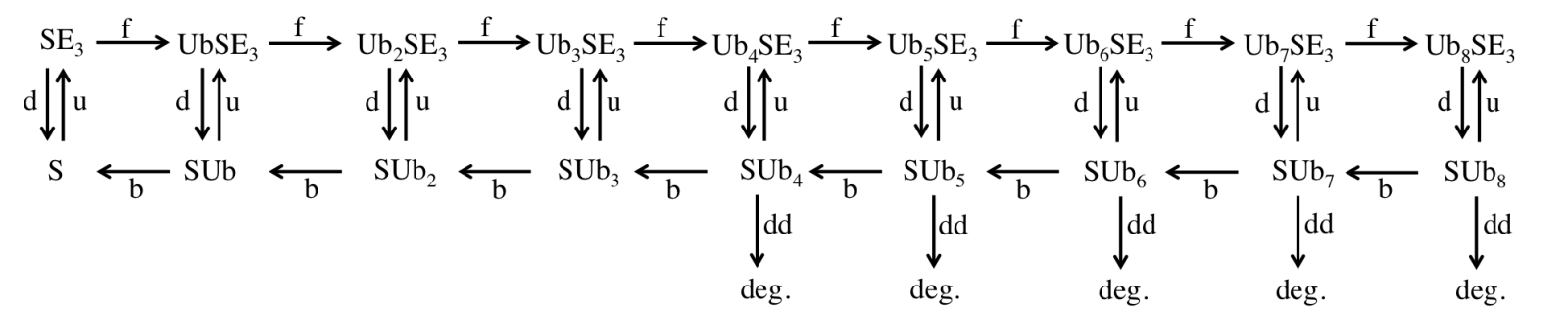
We will initially determine the number of fluorescent proteasomes in yeast. Then, using GFP that has been modified to include a recognition sequence for degradation, we will investigate this protein as its degradation is initiated by induction of an exogenous E3 and it is tagged and sent to the proteasome for removal. We will be able to measure diffusion constants and more since our STED can carry out in vivo fluorescence correlation spectroscopy (FCS) and can make nearly simultaneous FCS measurements with the multiple FCS spots that can be formed in a single sample. These measurements and further observation and analysis will further elucidate the spatio-temporal kinetics of the system.