Optical Tweezers
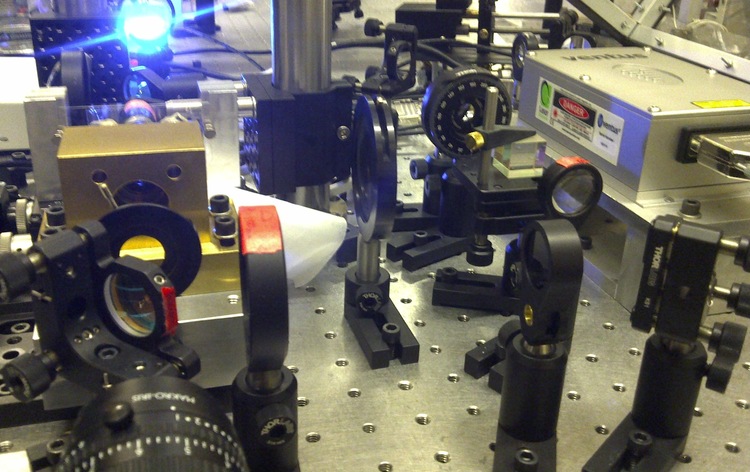
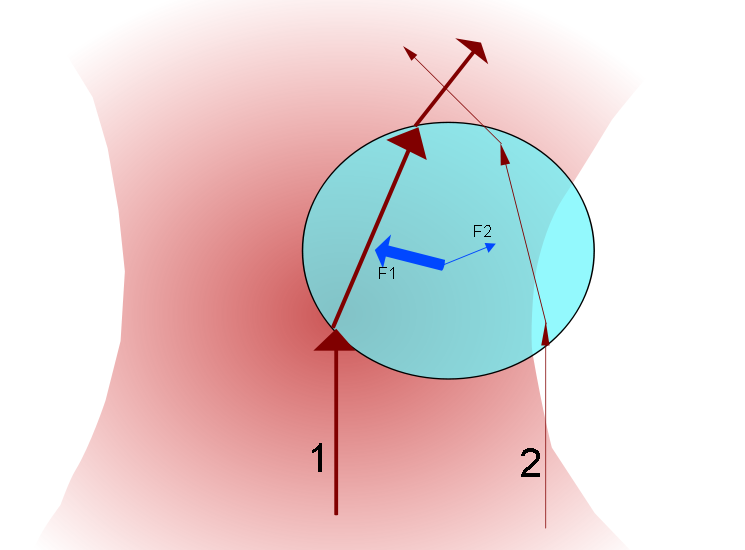
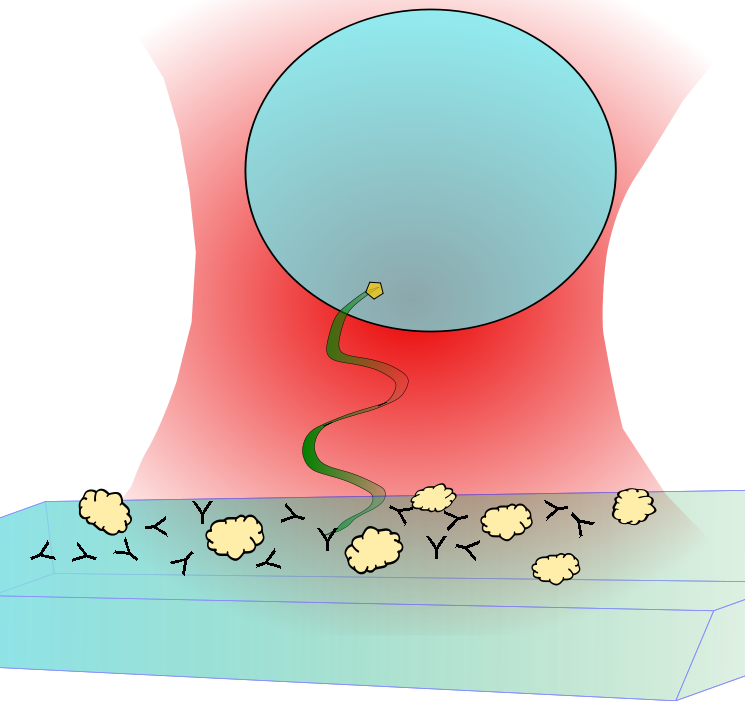
When a laser passes through a high numerical aperture objective (NA > 1.2), it creates a diffraction-limited focus with a steep intensity gradient. When a small spherical dielectric object (i.e. polystyrene or silica beads) with a high index of refraction is placed inside or near the focus, it gets trapped inside the harmonic potential of the focus. By appropriately labeling the ends of a DNA/RNA, we can exploit the surface chemistry to tether one end of the DNA/RNA to a cover slip and attach the other end to a bead. We can apply forces on the DNA/RNA constructs by either manipulating the position of the laser focus or the cover slip.
DNA Condensation
Every cell in your body has roughly one meter of DNA condensed into its one-micron nucleus. This is accomplished by special DNA packaging proteins, called histones. Packaging is highly regulated in order to package (turn off) and unpackage (turn on) genes along the DNA, as they are needed. Our initial studies have focused on measuring the force versus extension of DNA condensed by histone H1. Our measurements show remarkable agreement with theory, developed by us, describing DNA condensed into a helical toroid.
In addtion, using Crooks fluctuation theorem, we have developed a method to measure the energies of single molecule systems and state transition rates--from non-equilibrium measurements. We are using these methods to measure the binding energies of histones.
V(D)J Recombination
In all vertebrates (including humans), the immune system relies on V(D)J recombination to generate the diverse repertoire of all antigen receptors, most of which target invading pathogens. During the intiial stages of V(D)J recombination, RAG1 and RAG2 proteins catalyze double stranded DNA breaks at the junction between the coding segment and the neighboring Recombination Signal Sequences (RSS) in two steps. The first step of the reaction is just a nick of the upper single stranded DNA of this junction. The resulting 3'-hydroxl group at the coding end subsequently attacks the bottom DNA 5'phosphate strand at the RSS, forming a hairpin and a blunt end. The reaction occurs simultaneously at two RSSs paired by RAG combinase complex in the synapsis, while the intervening DNA is perhaps looped. We monitor the looping by tracking the Tethered Particle Motion and use Optical Tweezers to study the interactions of the DNA with the RAG proteins.